Analysis of hyperparameter search results#
In the previous notebook we showed how to implement a randomized search for
tuning the hyperparameters of a HistGradientBoostingClassifier to fit the
adult_census dataset. In practice, a randomized hyperparameter search is
usually run with a large number of iterations.
In order to avoid the computational cost and still make a decent analysis, we load the results obtained from a similar search with 500 iterations.
import pandas as pd
cv_results = pd.read_csv(
"../figures/randomized_search_results.csv", index_col=0
)
cv_results
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_classifier__l2_regularization | param_classifier__learning_rate | param_classifier__max_bins | param_classifier__max_leaf_nodes | param_classifier__min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.540456 | 0.062725 | 0.052069 | 0.002661 | 2.467047 | 0.550075 | 86 | 22 | 6 | {'classifier__l2_regularization': 2.4670474863... | 0.856558 | 0.862271 | 0.857767 | 0.854491 | 0.856675 | 0.857552 | 0.002586 | 48 |
| 1 | 1.110536 | 0.033403 | 0.074142 | 0.002165 | 0.015449 | 0.001146 | 60 | 19 | 1 | {'classifier__l2_regularization': 0.0154488709... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 2 | 1.137484 | 0.053150 | 0.092993 | 0.029005 | 1.095093 | 0.004274 | 151 | 17 | 10 | {'classifier__l2_regularization': 1.0950934559... | 0.783267 | 0.758941 | 0.776413 | 0.779143 | 0.758941 | 0.771341 | 0.010357 | 311 |
| 3 | 3.935108 | 0.202993 | 0.118105 | 0.023658 | 0.003621 | 0.001305 | 18 | 164 | 37 | {'classifier__l2_regularization': 0.0036210968... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 4 | 0.255219 | 0.038301 | 0.056048 | 0.016736 | 0.000081 | 5.407382 | 97 | 8 | 3 | {'classifier__l2_regularization': 8.1060737427... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 495 | 0.452411 | 0.023006 | 0.055563 | 0.000846 | 0.000075 | 0.364373 | 92 | 17 | 4 | {'classifier__l2_regularization': 7.4813767874... | 0.858332 | 0.865001 | 0.862681 | 0.860360 | 0.860770 | 0.861429 | 0.002258 | 34 |
| 496 | 1.133042 | 0.014456 | 0.078186 | 0.002199 | 5.065946 | 0.001222 | 7 | 17 | 1 | {'classifier__l2_regularization': 5.0659455480... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 497 | 0.911828 | 0.017167 | 0.076563 | 0.005130 | 2.460025 | 0.044408 | 16 | 7 | 7 | {'classifier__l2_regularization': 2.4600250010... | 0.839907 | 0.849713 | 0.846847 | 0.846028 | 0.844390 | 0.845377 | 0.003234 | 140 |
| 498 | 1.168120 | 0.121819 | 0.061283 | 0.000760 | 0.000068 | 0.287904 | 227 | 146 | 5 | {'classifier__l2_regularization': 6.7755366885... | 0.861881 | 0.865001 | 0.862408 | 0.859951 | 0.861862 | 0.862221 | 0.001623 | 33 |
| 499 | 0.823774 | 0.120686 | 0.060351 | 0.014958 | 0.445218 | 0.005112 | 19 | 8 | 19 | {'classifier__l2_regularization': 0.4452178932... | 0.764569 | 0.765902 | 0.765902 | 0.764947 | 0.765083 | 0.765281 | 0.000535 | 319 |
500 rows × 18 columns
We define a function to remove the prefixes in the hyperparameters column names.
def shorten_param(param_name):
if "__" in param_name:
return param_name.rsplit("__", 1)[1]
return param_name
cv_results = cv_results.rename(shorten_param, axis=1)
cv_results
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | l2_regularization | learning_rate | max_bins | max_leaf_nodes | min_samples_leaf | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.540456 | 0.062725 | 0.052069 | 0.002661 | 2.467047 | 0.550075 | 86 | 22 | 6 | {'classifier__l2_regularization': 2.4670474863... | 0.856558 | 0.862271 | 0.857767 | 0.854491 | 0.856675 | 0.857552 | 0.002586 | 48 |
| 1 | 1.110536 | 0.033403 | 0.074142 | 0.002165 | 0.015449 | 0.001146 | 60 | 19 | 1 | {'classifier__l2_regularization': 0.0154488709... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 2 | 1.137484 | 0.053150 | 0.092993 | 0.029005 | 1.095093 | 0.004274 | 151 | 17 | 10 | {'classifier__l2_regularization': 1.0950934559... | 0.783267 | 0.758941 | 0.776413 | 0.779143 | 0.758941 | 0.771341 | 0.010357 | 311 |
| 3 | 3.935108 | 0.202993 | 0.118105 | 0.023658 | 0.003621 | 0.001305 | 18 | 164 | 37 | {'classifier__l2_regularization': 0.0036210968... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 4 | 0.255219 | 0.038301 | 0.056048 | 0.016736 | 0.000081 | 5.407382 | 97 | 8 | 3 | {'classifier__l2_regularization': 8.1060737427... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 495 | 0.452411 | 0.023006 | 0.055563 | 0.000846 | 0.000075 | 0.364373 | 92 | 17 | 4 | {'classifier__l2_regularization': 7.4813767874... | 0.858332 | 0.865001 | 0.862681 | 0.860360 | 0.860770 | 0.861429 | 0.002258 | 34 |
| 496 | 1.133042 | 0.014456 | 0.078186 | 0.002199 | 5.065946 | 0.001222 | 7 | 17 | 1 | {'classifier__l2_regularization': 5.0659455480... | 0.758974 | 0.758941 | 0.758941 | 0.758941 | 0.758941 | 0.758947 | 0.000013 | 323 |
| 497 | 0.911828 | 0.017167 | 0.076563 | 0.005130 | 2.460025 | 0.044408 | 16 | 7 | 7 | {'classifier__l2_regularization': 2.4600250010... | 0.839907 | 0.849713 | 0.846847 | 0.846028 | 0.844390 | 0.845377 | 0.003234 | 140 |
| 498 | 1.168120 | 0.121819 | 0.061283 | 0.000760 | 0.000068 | 0.287904 | 227 | 146 | 5 | {'classifier__l2_regularization': 6.7755366885... | 0.861881 | 0.865001 | 0.862408 | 0.859951 | 0.861862 | 0.862221 | 0.001623 | 33 |
| 499 | 0.823774 | 0.120686 | 0.060351 | 0.014958 | 0.445218 | 0.005112 | 19 | 8 | 19 | {'classifier__l2_regularization': 0.4452178932... | 0.764569 | 0.765902 | 0.765902 | 0.764947 | 0.765083 | 0.765281 | 0.000535 | 319 |
500 rows × 18 columns
As we have more than 2 parameters in our randomized-search, we cannot visualize the results using a heatmap. We could still do it pair-wise, but having a two-dimensional projection of a multi-dimensional problem can lead to a wrong interpretation of the scores.
import seaborn as sns
import numpy as np
df = pd.DataFrame(
{
"max_leaf_nodes": cv_results["max_leaf_nodes"],
"learning_rate": cv_results["learning_rate"],
"score_bin": pd.cut(
cv_results["mean_test_score"], bins=np.linspace(0.5, 1.0, 6)
),
}
)
sns.set_palette("YlGnBu_r")
ax = sns.scatterplot(
data=df,
x="max_leaf_nodes",
y="learning_rate",
hue="score_bin",
s=50,
color="k",
edgecolor=None,
)
ax.set_xscale("log")
ax.set_yscale("log")
_ = ax.legend(
title="mean_test_score", loc="center left", bbox_to_anchor=(1, 0.5)
)
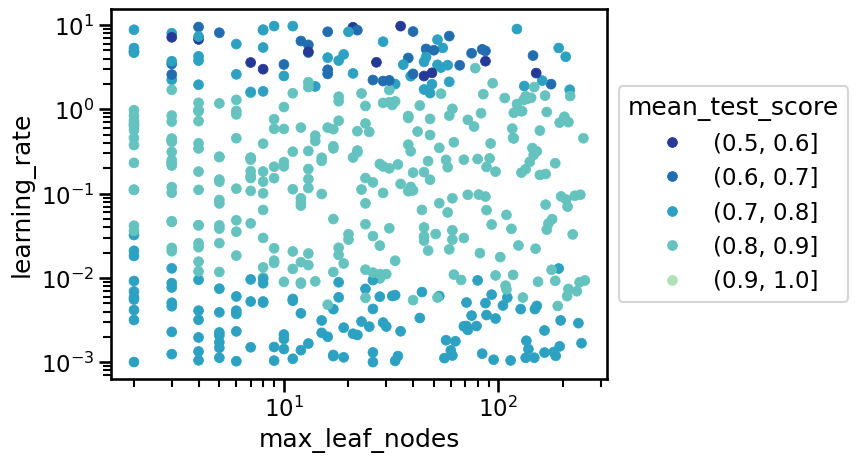
In the previous plot we see that the top performing values are located in a band of learning rate between 0.01 and 1.0, but we have no control in how the other hyperparameters interact with such values for the learning rate. Instead, we can visualize all the hyperparameters at the same time using a parallel coordinates plot.
import numpy as np
import plotly.express as px
fig = px.parallel_coordinates(
cv_results.rename(shorten_param, axis=1).apply(
{
"learning_rate": np.log10,
"max_leaf_nodes": np.log2,
"max_bins": np.log2,
"min_samples_leaf": np.log10,
"l2_regularization": np.log10,
"mean_test_score": lambda x: x,
}
),
color="mean_test_score",
color_continuous_scale=px.colors.sequential.Viridis,
)
fig.show(renderer="notebook")
Note
We transformed most axis values by taking a log10 or log2 to spread the active ranges and improve the readability of the plot.
The parallel coordinates plot displays the values of the hyperparameters on different columns while the performance metric is color coded. Thus, we are able to quickly inspect if there is a range of hyperparameters which is working or not.
It is possible to select a range of results by clicking and holding on any axis of the parallel coordinate plot. You can then slide (move) the range selection and cross two selections to see the intersections. You can undo a selection by clicking once again on the same axis.
In particular for this hyperparameter search, it is interesting to confirm that the yellow lines (top performing models) all reach intermediate values for the learning rate, that is, tick values between -2 and 0 which correspond to learning rate values of 0.01 to 1.0 once we invert back the log10 transform for that axis.
But now we can also observe that it is not possible to select the highest
performing models by selecting lines of on the max_bins axis with tick
values between 1 and 3.
The other hyperparameters are not very sensitive. We can check that if we
select the learning_rate axis tick values between -1.5 and -0.5 and
max_bins tick values between 5 and 8, we always select top performing
models, whatever the values of the other hyperparameters.
In this notebook, we saw how to interactively explore the results of a large randomized search with multiple interacting hyperparameters. In particular we observed that some hyperparameters have very little impact on the cross-validation score, while others have to be adjusted within a specific range to get models with good predictive accuracy.

