📃 Solution for Exercise M7.02#
We presented different classification metrics in the previous notebook. However, we did not use it with a cross-validation. This exercise aims at practicing and implementing cross-validation.
Here we use the blood transfusion dataset.
import pandas as pd
blood_transfusion = pd.read_csv("../datasets/blood_transfusion.csv")
data = blood_transfusion.drop(columns="Class")
target = blood_transfusion["Class"]
Note
If you want a deeper overview regarding this dataset, you can refer to the Appendix - Datasets description section at the end of this MOOC.
First, create a decision tree classifier.
# solution
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier()
Create a StratifiedKFold cross-validation object. Then use it inside the
cross_val_score function to evaluate the decision tree. We first use
the accuracy as a score function. Explicitly use the scoring parameter of
cross_val_score to compute the accuracy (even if this is the default score).
Check its documentation to learn how to do that.
# solution
from sklearn.model_selection import cross_val_score, StratifiedKFold
cv = StratifiedKFold(n_splits=10)
scores = cross_val_score(tree, data, target, cv=cv, scoring="accuracy")
print(f"Accuracy score: {scores.mean():.3f} ± {scores.std():.3f}")
Accuracy score: 0.623 ± 0.144
Repeat the experiment by computing the balanced_accuracy.
# solution
scores = cross_val_score(
tree, data, target, cv=cv, scoring="balanced_accuracy"
)
print(f"Balanced accuracy score: {scores.mean():.3f} ± {scores.std():.3f}")
Balanced accuracy score: 0.512 ± 0.105
We now add a bit of complexity. We would like to compute the precision of
our model. However, during the course we saw that we need to mention the
positive label which in our case we consider to be the class donated.
We can show that computing the precision without providing the positive label is not supported by scikit-learn because it is indeed ambiguous.
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier()
try:
scores = cross_val_score(
tree, data, target, cv=10, scoring="precision", error_score="raise"
)
except ValueError as exc:
print(exc)
pos_label=1 is not a valid label: It should be one of ['donated' 'not donated']
Tip
We use a try/except block to catch possible ValueErrors and print them
if they occur. By setting error_score="raise", we ensure that the exception
is raised immediately when an error is encountered. Without this setting, the
code would show a warning for each cross-validation split before raising the
exception. You can try using the default error_score to better understand
what this means.
We get an exception because the default scorer has its positive label set to
one (pos_label=1), which is not our case (our positive label is “donated”).
In this case, we need to create a scorer using the scoring function and the
helper function make_scorer.
So, import sklearn.metrics.make_scorer and
sklearn.metrics.precision_score. Check their documentations for more
information. Finally, create a scorer by calling make_scorer using the score
function precision_score and pass the extra parameter pos_label="donated".
# solution
from sklearn.metrics import make_scorer, precision_score
precision = make_scorer(precision_score, pos_label="donated")
Now, instead of providing the string "precision" to the scoring parameter
in the cross_val_score call, pass the scorer that you created above.
# solution
scores = cross_val_score(tree, data, target, cv=cv, scoring=precision)
print(f"Precision score: {scores.mean():.3f} ± {scores.std():.3f}")
Precision score: 0.242 ± 0.174
cross_val_score can compute one score at a time, as specified by the scoring
parameter. In contrast, cross_validate can compute multiple scores by passing
a list of strings or scorers to the scoring parameter
Import sklearn.model_selection.cross_validate and compute the accuracy and
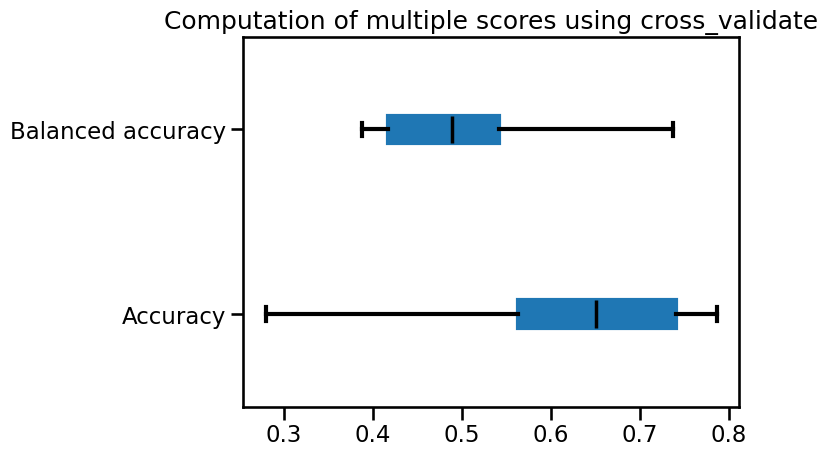
balanced accuracy through cross-validation. Plot the cross-validation score
for both metrics using a box plot.
# solution
from sklearn.model_selection import cross_validate
scoring = ["accuracy", "balanced_accuracy"]
scores = cross_validate(tree, data, target, cv=cv, scoring=scoring)
scores
{'fit_time': array([0.00290465, 0.00274086, 0.0027554 , 0.00275826, 0.00277567,
0.00276637, 0.00268245, 0.00274873, 0.00273824, 0.00271893]),
'score_time': array([0.00270462, 0.00259829, 0.00251675, 0.00257778, 0.00260758,
0.00264812, 0.0026226 , 0.00262618, 0.00263548, 0.00266218]),
'test_accuracy': array([0.28 , 0.50666667, 0.78666667, 0.57333333, 0.56 ,
0.64 , 0.69333333, 0.77333333, 0.66216216, 0.75675676]),
'test_balanced_accuracy': array([0.39327485, 0.46637427, 0.68859649, 0.41520468, 0.3874269 ,
0.42105263, 0.55116959, 0.73684211, 0.5123839 , 0.51186791])}
import pandas as pd
color = {"whiskers": "black", "medians": "black", "caps": "black"}
metrics = pd.DataFrame(
[scores["test_accuracy"], scores["test_balanced_accuracy"]],
index=["Accuracy", "Balanced accuracy"],
).T
import matplotlib.pyplot as plt
metrics.plot.box(vert=False, color=color)
_ = plt.title("Computation of multiple scores using cross_validate")